From Average to Highly Specialized: A Beginner guide to Single-Cell RNA-Seq (scRNA-seq)
Part1:
Welcome to the first blog in our series on single-cell RNA sequencing (scRNA-seq). In this series, we will explore the fascinating world of scRNA-seq, from its basics to advanced analysis techniques. Whether you’re a beginner or looking to deepen your understanding, this series is designed to guide you through the complexities of scRNA-seq.
Cell being the building block of life:
Life, as we know it, is the characteristics that differentiate living from non-living things. Almost all form of life on earth shares a similar identity at microscopic level that we called, Cells. I am sure that all of you would have studied the famous Cell Theory in your School (with some modern interpretation) that says:
- All living organisms are composed of one or more cells.
- Cell is the basic unit of structure and organization in organisms.
- All cells arise from pre-existing cells.
- The activity of an organism depends on the total activity of independent cells.
- Energy flow (metabolism and biochemistry) occurs within cells.
- Cells contain DNA which is found specifically in the chromosome and RNA found in the cell nucleus and cytoplasm.
- All cells are basically the same in chemical composition in organisms of similar species.
Why Single Cell ?
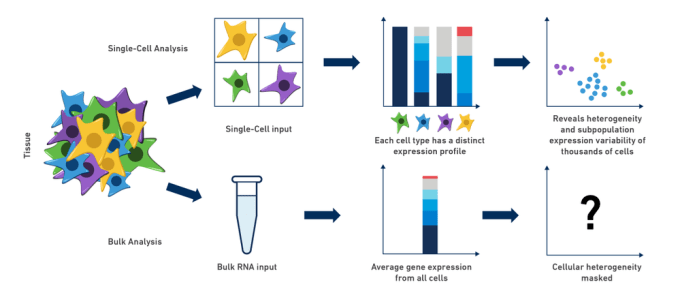
Complex Biology give rise to complex system, where each component of the system works in a coordinated fashion to their own special role in order to be as important as others. Before the advent of single-cell techniques we were only able to understand and analyze cell-averaged expression profile which was good but were unable to answer certain questions:
- cell-to-cell differences were missing.
- cellular heterogeneity completely masked.
Now with single cell one can draw a comparison not only at inter-tumor but at intra-tumor level which was not possible in case of Bulk-rnaseq. Scrna-seq based studies can be used to:
Explore which cell types are present in a tissue (Cellular Heterogeneity)
Identify unknown/rare cell types or states
Elucidate the changes in gene expression during differentiation processes or across time or states
Identify genes that are differentially expressed in particular cell types between conditions (e.g. treatment or disease or treated vs untreated)
Explore changes in expression among a cell type while incorporating spatial, regulatory, and/or protein information
Challenges with scRNA-seq Analysis:
Bulk RNA-seq works perfectly well if your analysis involves comparing average-cellular expression between two different conditions. Similarly it also has potential for the discovery of disease biomarkers if you are not expecting or not concerned about cellular heterogeneity in the sample. However, scrna-seq with its revolutionizing capabilities to uncover cellular heterogeneity also has some challenges associated with it.
Experiment is costly.
Library preparation is difficult.
Analysis of the data is complicated.
Difficult to interpret data compare to bulk rna-seq analysis.
Large volume of data
Low depth of sequencing per cell
Technical variability across cells/samples
Biological variability across cells/samples
More on these challenges in the next blog post which will be part 2 of this series.